-Search query
-Search result
Showing 1 - 50 of 85 items for (author: sanchez & rm)

EMDB-42464: 
chEnv TTT protein in complex with 43A2 Fab
Method: single particle / : Ozorowski G, Lee WH, Ward AB

EMDB-42468: 
chEnv TTT protein in complex with CM01A Fab
Method: single particle / : Ozorowski G, Lee WH, Ward AB

EMDB-43664: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines
Method: single particle / : Ferguson JA, Leon AN, Ward AB

EMDB-43665: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (cH125 TTT)
Method: single particle / : Ferguson JA, Leon AN, Ward AB

EMDB-43666: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (H2/1 GCN4)
Method: single particle / : Ferguson JA, Leon AN, Ward AB

EMDB-43668: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines (H5/1 GCN4)
Method: single particle / : Ferguson JA, Leon AN, Ward AB

EMDB-43669: 
Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines. H5 GCN4
Method: single particle / : Ferguson JA, Leon AN, Ward AB

EMDB-19250: 
Pseudoatomic model of a second-order Sierpinski triangle formed by the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-19251: 
Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
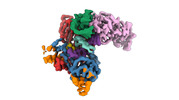
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
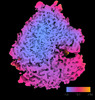
EMDB-18505: 
Human 80S ribosome structure from pFIB-lamellae reprocessed with TomoBEAR
Method: subtomogram averaging / : Balyschew N, Yushkevich A, Mikirtumov V, Sanchez RM, Sprink T, Kudryashev M

EMDB-40789: 
BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI, Armache KJ

EMDB-40790: 
Map focused on acidic patch BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI

EMDB-40791: 
Overall map of BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI

PDB-8svf: 
BAP1/ASXL1 bound to the H2AK119Ub Nucleosome
Method: single particle / : Thomas JF, Valencia-Sanchez MI, Armache KJ

EMDB-17272: 
Structure of RyR1 obtained from SR vesicles tilt series (EMPIAR-10452), reprocessed with TomoBEAR
Method: subtomogram averaging / : Balyschew N, Yushkevich A, Mikirtumov V, Sanchez RM, Sprink T, Kudryashev M
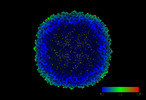
EMDB-17232: 
Purified human apoferritin at 2.8 A resolution from tilt series processed with TomoBEAR
Method: subtomogram averaging / : Balyschew N, Yushkevich A, Mikirtumov V, Sanchez RM, Sprink T, Kudryashev M
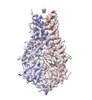
EMDB-28538: 
Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

EMDB-28544: 
Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

EMDB-28545: 
Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE
EMDB-28546: 
Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

PDB-8eqj: 
Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

PDB-8eqs: 
Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

PDB-8eqt: 
Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

PDB-8equ: 
Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc
Method: single particle / : Miller AN, Houlihan PR, Matamala E, Cabezas-Bratesco D, Lee GY, Cristofori-Armstrong B, Dilan TL, Sanchez-Martinez S, Matthies D, Yan R, Yu Z, Ren D, Brauchi SE, Clapham DE

EMDB-13916: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Frances-Gomez C, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, LLacer JL, Carazo JM

EMDB-13917: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 2-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM

EMDB-13918: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 3-down
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM

EMDB-13919: 
SARS-CoV-2 S protein S:D614G mutant 1-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Frances-Gomez C, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, LLacer JL, Carazo JM
EMDB-13920: 
SARS-CoV-2 S protein S:D614G mutant 2-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, Llacer JL, Carazo JM

PDB-7qdg: 
SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Frances-Gomez C, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, LLacer JL, Carazo JM

PDB-7qdh: 
SARS-CoV-2 S protein S:D614G mutant 1-up
Method: single particle / : Ginex T, Marco-Marin C, Wieczor M, Mata CP, Krieger J, Lopez-Redondo ML, Frances-Gomez C, Ruiz-Rodriguez P, Melero R, Sanchez-Sorzano CO, Martinez M, Gougeard N, Forcada-Nadal A, Zamora-Caballero S, Gozalbo-Rovira R, Sanz-Frasquet C, Bravo J, Rubio V, Marina A, Geller R, Comas I, Gil C, Coscolla M, Orozco M, LLacer JL, Carazo JM

EMDB-13153: 
2.43 A Mycobacterium marinum EspB.
Method: single particle / : Gijsbers A, Zhang Y, Vinciauskaite V, Siroy A, Ye G, Tria G, Mathew A, Sanchez-Puig N, Lopez-Iglesias C, Peters PJ, Ravelli RBG

EMDB-13154: 
2.29 A Mycobacterium tuberculosis EspB.
Method: single particle / : Gijsbers A, Zhang Y, Vinciauskaite V, Siroy A, Gao Y, Tria G, Mathew A, Sanchez-Puig N, Lopez-Iglesias C, Peters PJ, Ravelli RBG

PDB-7p0z: 
2.43 A Mycobacterium marinum EspB.
Method: single particle / : Gijsbers A, Zhang Y, Vinciauskaite V, Siroy A, Ye G, Tria G, Mathew A, Sanchez-Puig N, Lopez-Iglesias C, Peters PJ, Ravelli RBG

PDB-7p13: 
2.29 A Mycobacterium tuberculosis EspB.
Method: single particle / : Gijsbers A, Zhang Y, Vinciauskaite V, Siroy A, Gao Y, Tria G, Mathew A, Sanchez-Puig N, Lopez-Iglesias C, Peters PJ, Ravelli RBG

EMDB-11860: 
Subtomogram average structure of anammoxosomal nitrite oxidoreductase
Method: subtomogram averaging / : Dietrich L

EMDB-11861: 
Helical reconstruction of nitrite oxidoreductase from anammox bacteria
Method: helical / : Parey K

EMDB-23529: 
Structure of the Marseillevirus nucleosome
Method: single particle / : Valencia-Sanchez MI, Abini-Agbomson S, Armache KJ

EMDB-23530: 
Marseillevirus heterotrimeric (hexameric) nucleosome
Method: single particle / : Valencia-Sanchez MI, Abini-Agbomson S, Armache KJ

PDB-7lv8: 
Structure of the Marseillevirus nucleosome
Method: single particle / : Valencia-Sanchez MI, Abini-Agbomson S, Armache KJ

PDB-7lv9: 
Marseillevirus heterotrimeric (hexameric) nucleosome
Method: single particle / : Valencia-Sanchez MI, Abini-Agbomson S, Armache KJ

EMDB-22691: 
Active state Dot1 bound to the unacetylated H4 nucleosome
Method: single particle / : Valencia-Sanchez MI, De Ioannes PE

EMDB-22692: 
Active state Dot1 bound to the H4K16ac nucleosome
Method: single particle / : Valencia-Sanchez MI, De Ioannes PE, Miao W, Truong DM, Lee R, Armache JP, Boeke JD, Armache KJ

EMDB-22693: 
Noncatalytic conformation Dot1 bound to the unacetylated H4 nucleosome
Method: single particle / : Valencia-Sanchez MI, De Ioannes PE, Miao W, Truong DM, Lee R, Armache JP, Boeke JD, Armache KJ

EMDB-22694: 
Active state Dot1 bound to the H4K16ac nucleosome, Ubiquitin focused map
Method: single particle / : Valencia-Sanchez MI, De Ioannes PE, Miao W, Truong DM, Lee R, Armache JP, Boeke JD, Armache KJ

EMDB-22695: 
Active state Dot1 bound to the unacetylated H4 nucleosome, Ubiquitin focused map
Method: single particle / : Valencia-Sanchez MI, De Ioannes PE, Miao W, Truong DM, Lee R, Armache JP, Boeke JD, Armache KJ

PDB-7k6p: 
Active state Dot1 bound to the unacetylated H4 nucleosome
Method: single particle / : Valencia-Sanchez MI, De Ioannes PE, Miao W, Truong DM, Lee R, Armache JP, Boeke JD, Armache KJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
